Ramlan explains his bacterial grid display model and how it fits into the larger picture”Loss works on any” tradition.
At Ars, we’ve covered versions of Loss running on everything from hacked printers to Windows notepad.exe to a version running inside Loss himself. But these and other numerous and varied examples of oddities Loss platforms not all of them have the pure biological quirk of a new game display model using a grid of E.coli bacteria.
MIT graduate student Lauren Ramlan describes a method for creating the Quixote Loss show in “1-bit pixels encoded in E. coli for interactive digital media display”, the final project of a Principles of synthetic biology class. The Ramlan project is based on previous research describing how DNA in E.coli bacteria can be used to encode fully digital circuits and how bacteria can be made to fluoresce as a crude form of digital display.
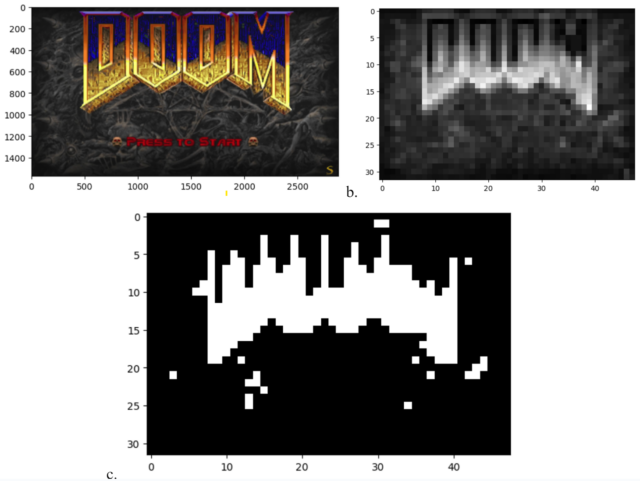
Ramlan’s article does not go to the enormous trouble of coding the entire Loss to function in bacterial DNA, which the author describes as “a colossal feat that I can’t even imagine approaching.” Instead, the game runs on a standard computer, with E.coli cells in a standard 32×48 microwell grid serving as a low-resolution raw display.
After reducing each frame in the game to a 32×48 black and white bitmap, Ramlan describes a system whereby a display controller uses a well-known chemical repressor-operator pair to induce each individual cell in the grid to express or not express a fluorescent protein. The resulting grid of luminous bacteria (which is only simulated in Ramlan’s project) can technically be considered a display of Loss gameplay, although the lack of shading, even in grayscale, makes the resulting image quite indecipherable, to be honest.
The time required for this entire process also limits the usefulness of the theoretical bacterial display. Using a Python model, Ramlan calculates that it would take 8 hours and 20 minutes to get a glowing light. E.coli cell to “return approximately to its starting state”, including a full 70 minutes to “reach maximum display output” during the cycle. This translates to a frame rate of 0.00003 fps which would span five hours Loss come up against a work of 599 years more suitable for Total attention span than the length of human life.
All of these caveats combine to make this one of the most technical additions to the ever growing “Loss works on all” list. Still, this effort shows just how far some people are willing to go to turn anything into a theoretical theory. Loss-display ready. As Ramlan writes in his article: “To execute Lossall you need is a screen and willpower.”
Listing image by parfkfkrErbe / Christopher Pooley